
Transform BAM files into fastq format.īam_stat.py: Now counts ‘Proper-paired reads map to different chrom’īam2wig.py: automatically call ‘wigToBigwig’ if it can be found in system $PATH Generate heatmap to visualize gene body coverage over many samples.Īdd bam2fq.py. Input a directory containing BAM file ( s ). Input plain text file containing the path of BAM file ( s ). Input several BAM files ( separated by "," ). It does not report exon and intron level count.ġ. FPKM.py will report “raw fragment count”, “FPM” and “FPKM” for each gene. This happened when reads were clipped and spliced mapped simultaneously.Īdd FPKM.py. “bam_stat.py” prints summary statistics to STDOUT.įix bugs in “insertion_profile.py”, “clipping_profile.py”, and “inner_distance.py “įix bug in “junction_annotation.py” in that it would report some “novel splice junctions” that don’t exist in the BAM files. Remove “RPKM_count.py” as it generates erroneous results especially for longer reads.
bx-python and pysam will be installed automatically if they haven’t been installed before.įix a bug in “read_quality.py” that does not return results if input file containing less than 1000 reads.
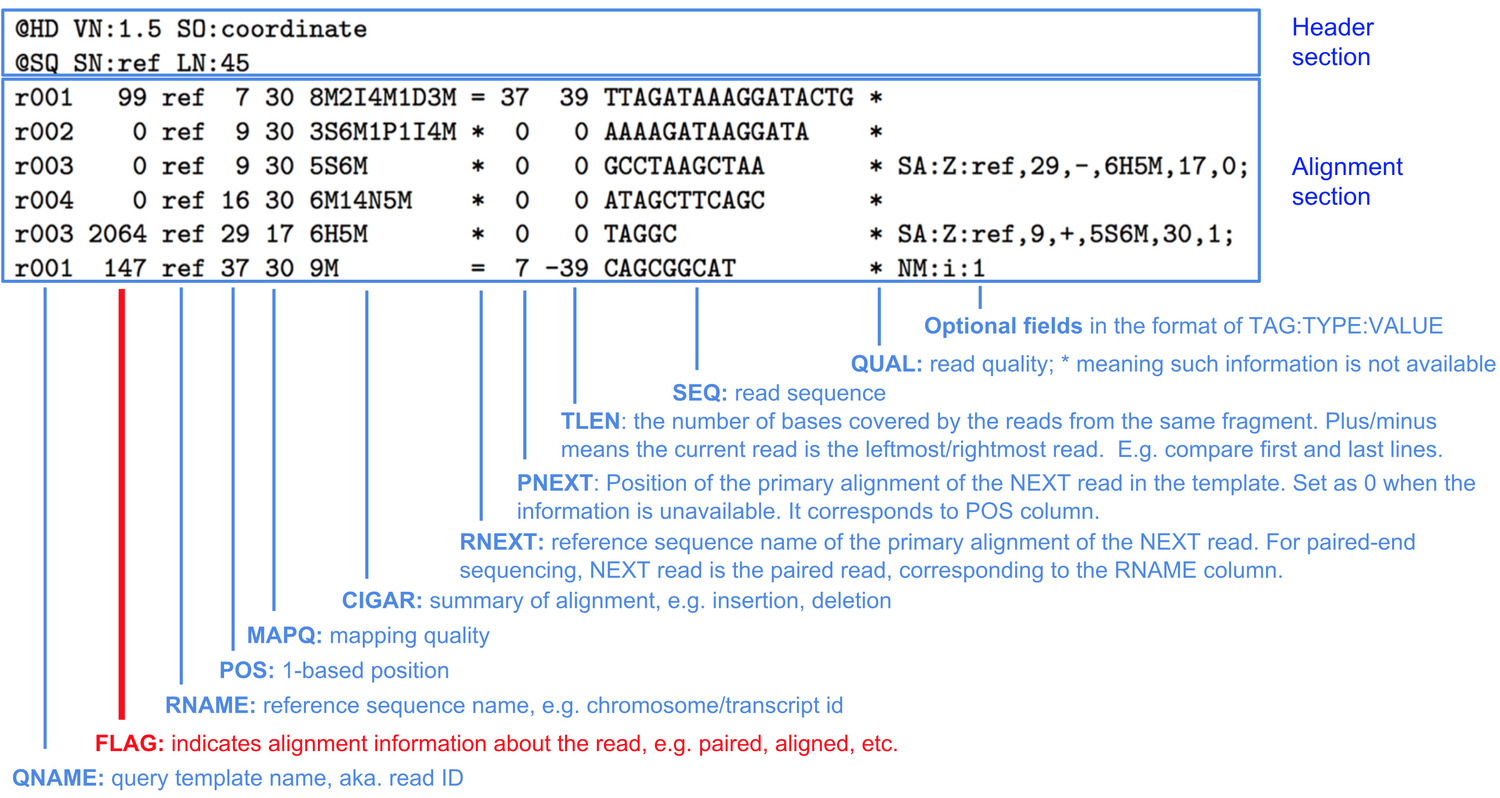
Bam file format columns install#
Users could install RSeQC using pip: pip install RSeQC. Two dependency packages bx-python and pysam are not shipped with RSeQC starting from v2.6.4. Please use previous versions (v2.6.5 or older) if you are using Python2. Junctions detected from the junction_annotation.py will be converted into Interact format file, which can be uploaded into UCSC genome browser for visualization. Further information is available on the FTP site.Add FPKM-UQ.py to calcualte HTSeq count, FPKM and FPKM-UQ values defined by TCGAįPKM-UQ.py could exactly reproduce TCGA FPKM-UQ values, if you use TCGA BAM file (or follow TCGA RNA-seq alignment workflow to generate your own BAM file), the GDC.h38 GENCODE v22 GTF file and the GDC.h38 GENCODE TSV file. These lines start with # and can provide descriptions of the columns, the date the index was generated and other pieces of information, as appropriate to the file and data set. In addition, index files may have further information at the head of the file. Immediately before the body of the file there is a header line, which starts with #, that gives the column names. The index files are tab-delimited files where the data is arranged in columns. Various types of index file exist on the site, primarily listing available sequence data and alignments. Further information is available on the FTP site.

Result for a column, the default value will be 0. Where data isn’t available to calculate the Provide meta information about each readgroup, with the remaining columns providing various statistics about the readgroup. The first line is a header that describes each column. cram files, with one line per readgroup and columns separated by bas files contain statistics relating to. Current specifications for SAM/BAM, CRAM and VCF can be found at hts-specs. The specifications for these file formats continue to develop. It is able to store all variant calls from single nucleotide variants to large scale insertions and deletions. The VCF format is a tab delimited format for storing variant calls and individual genotypes.

Information on working with IGSR CRAM files are available on the FTP site. The file format was designed to reduce the disk foot print of alignment data by the EBI, who provide further information on the format. This compression is driven by the reference the sequence data is aligned to. ĬRAM files are similar to BAM files but give a compressed representation of the alignment. These files and the associated SAMtools package are described in this Bioinformatics publication.Īdditional information about SAM/BAM is available at the SAMtools development site. Data file formats Alignment files: BAM and CRAM formatsīAM files are binary representations of the Sequence Alignment/Map format.


 0 kommentar(er)
0 kommentar(er)
